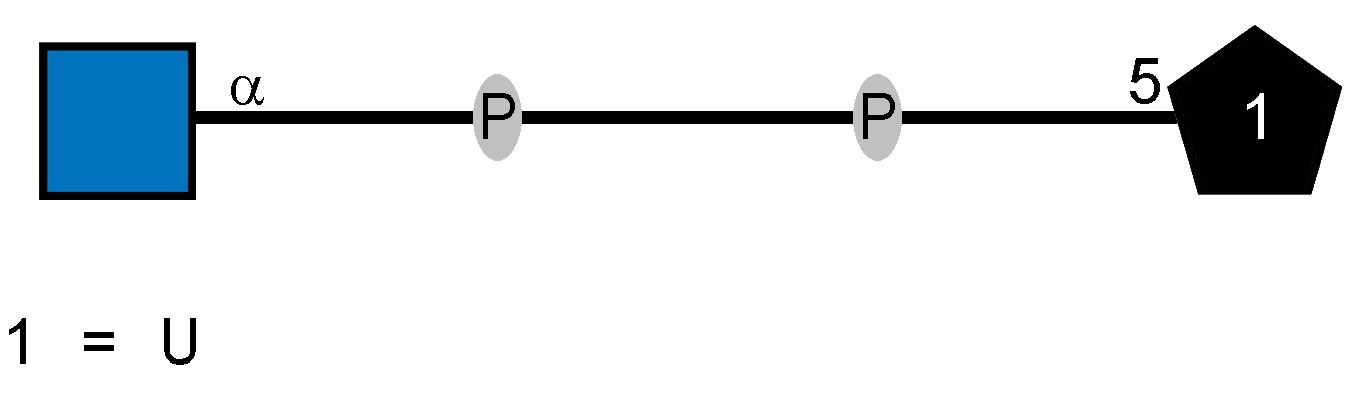
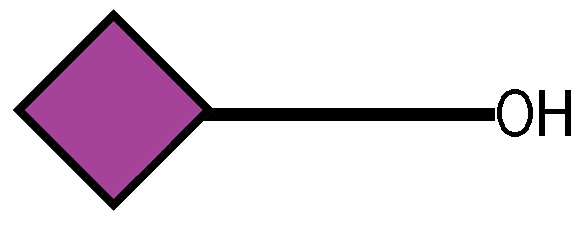
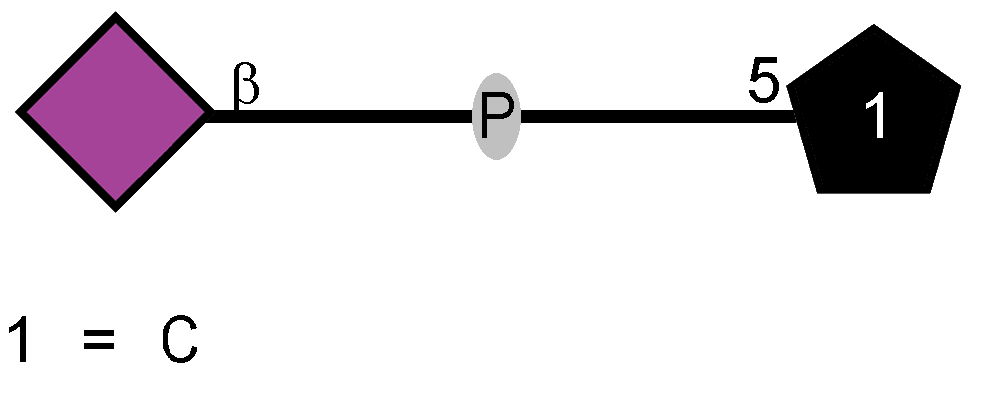
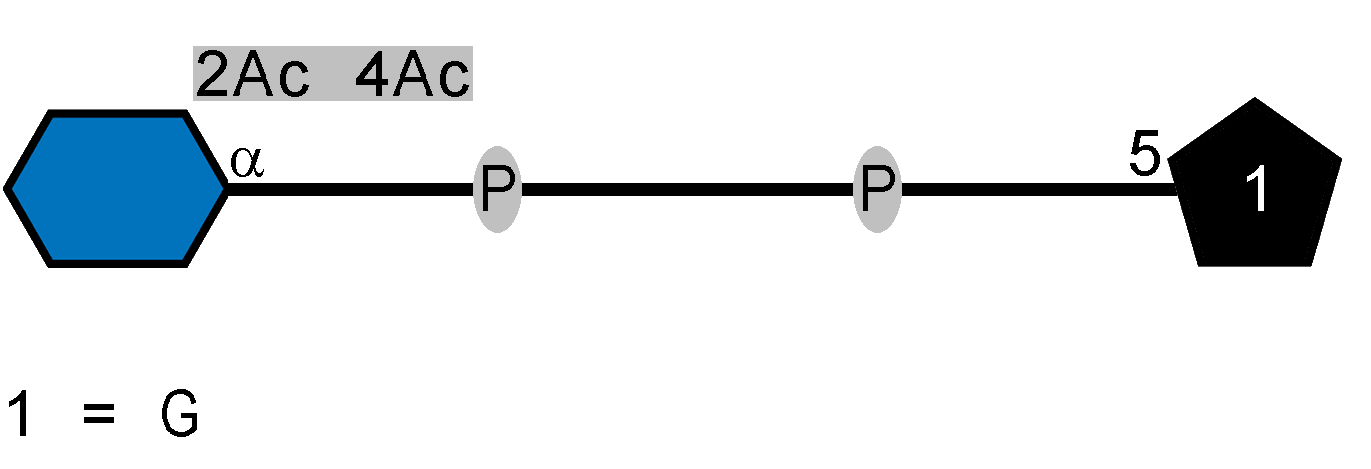
Genes
Glycans
All data
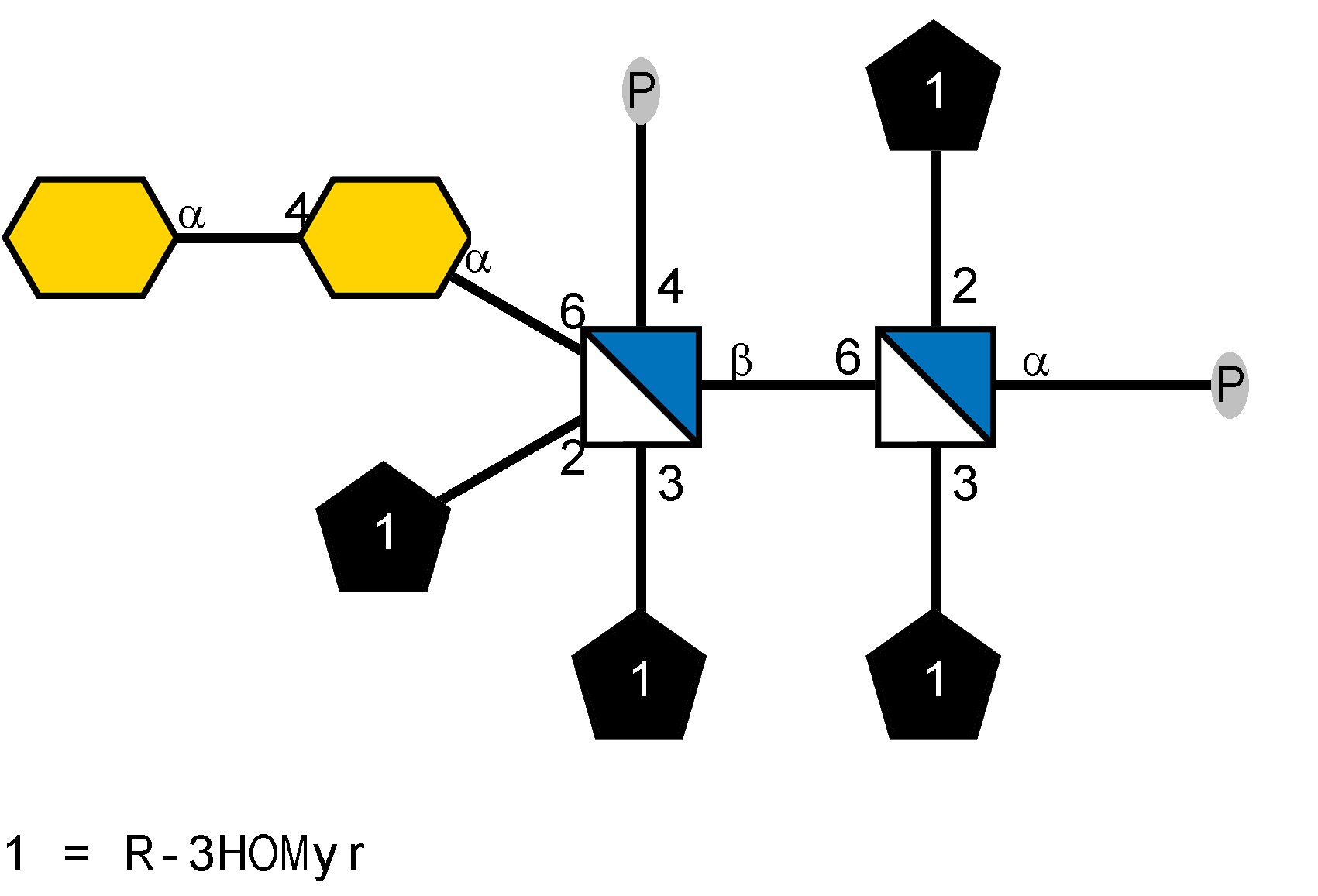
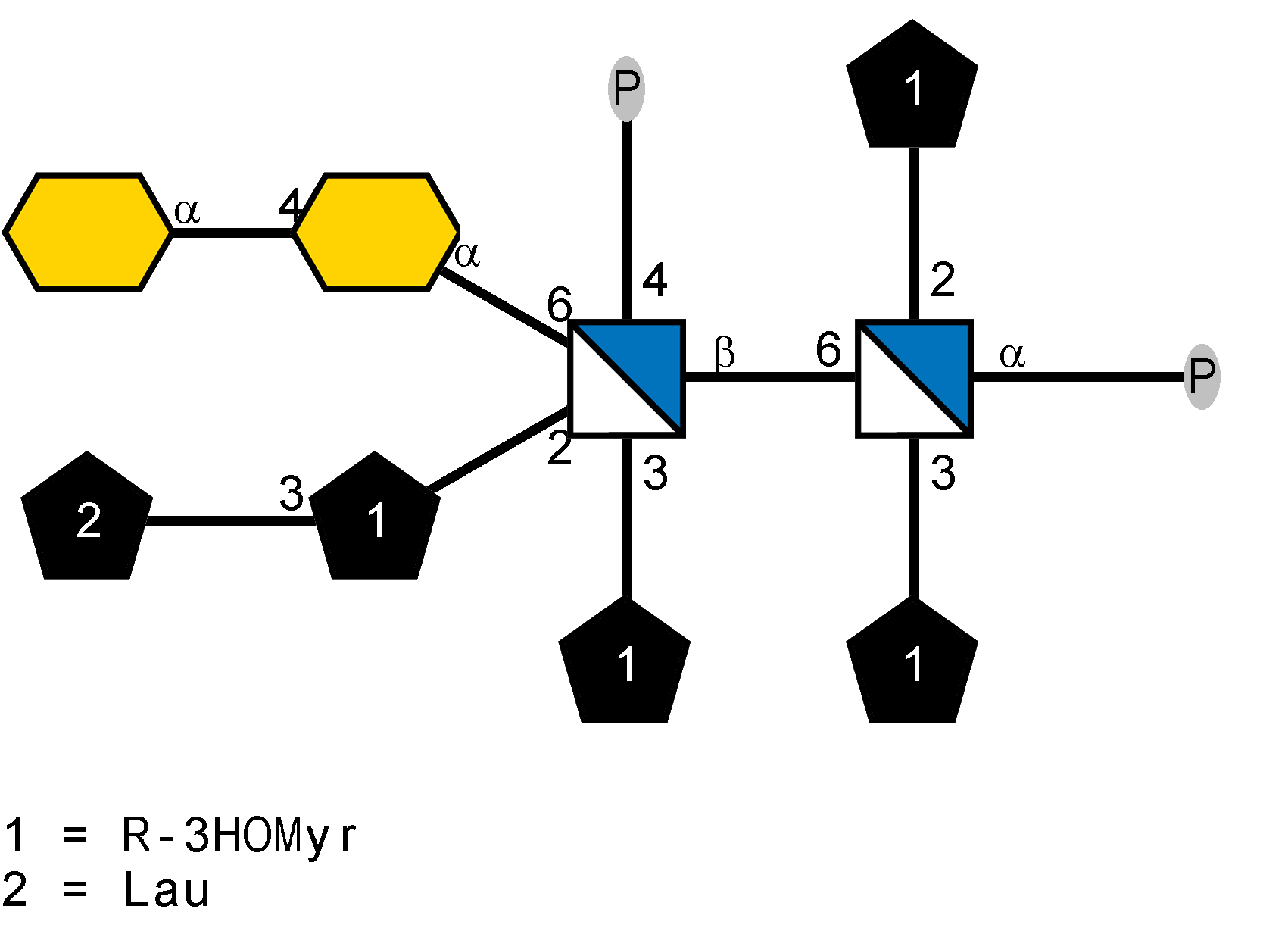
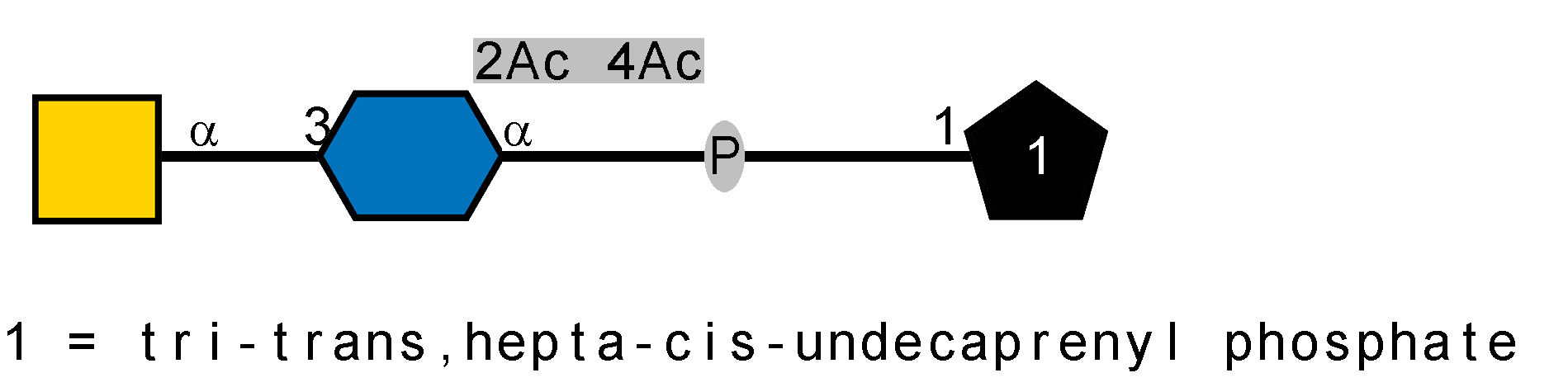
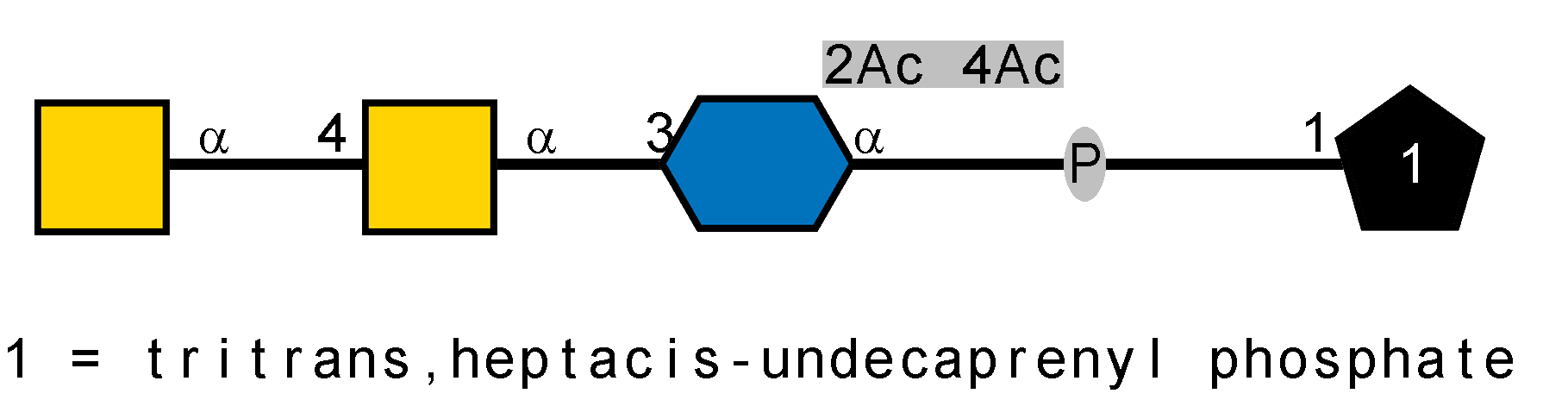
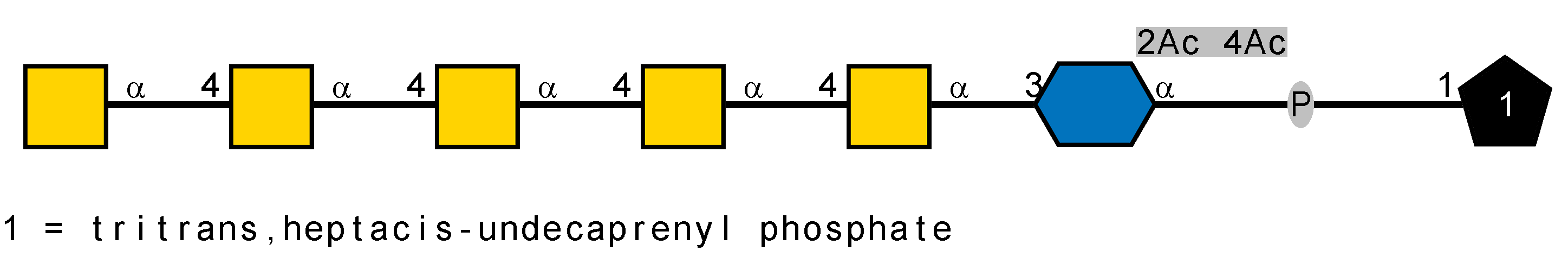
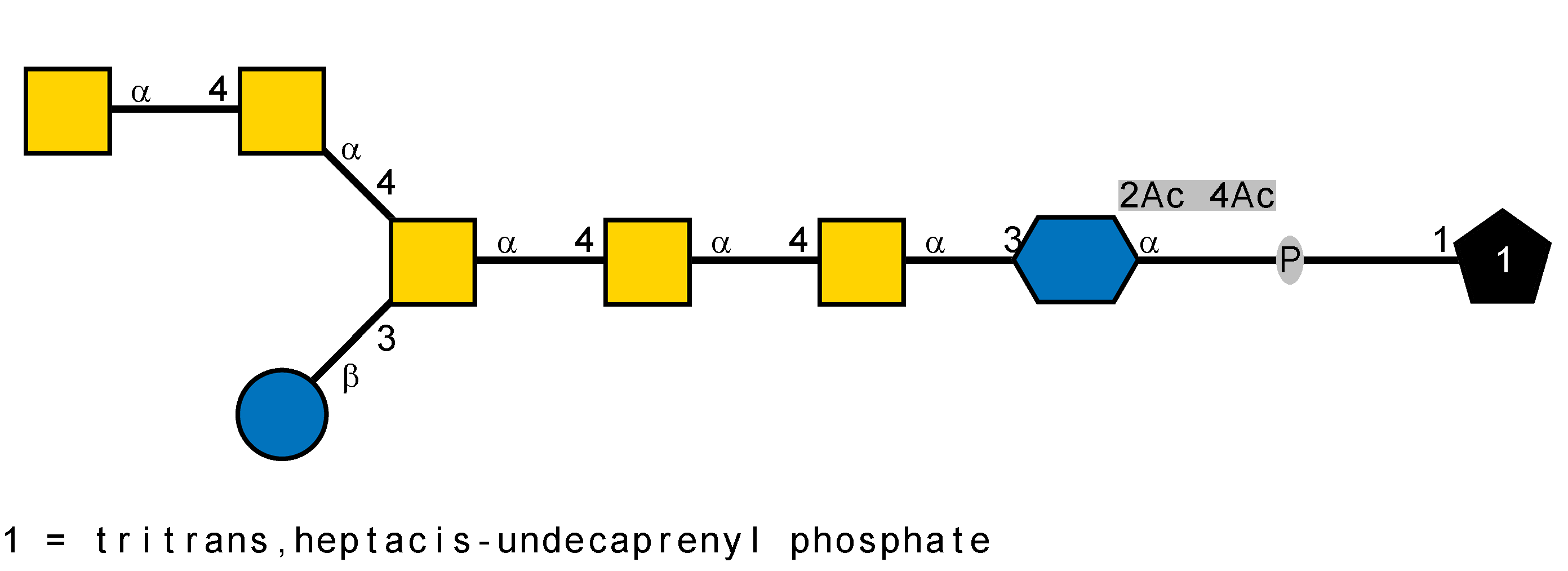
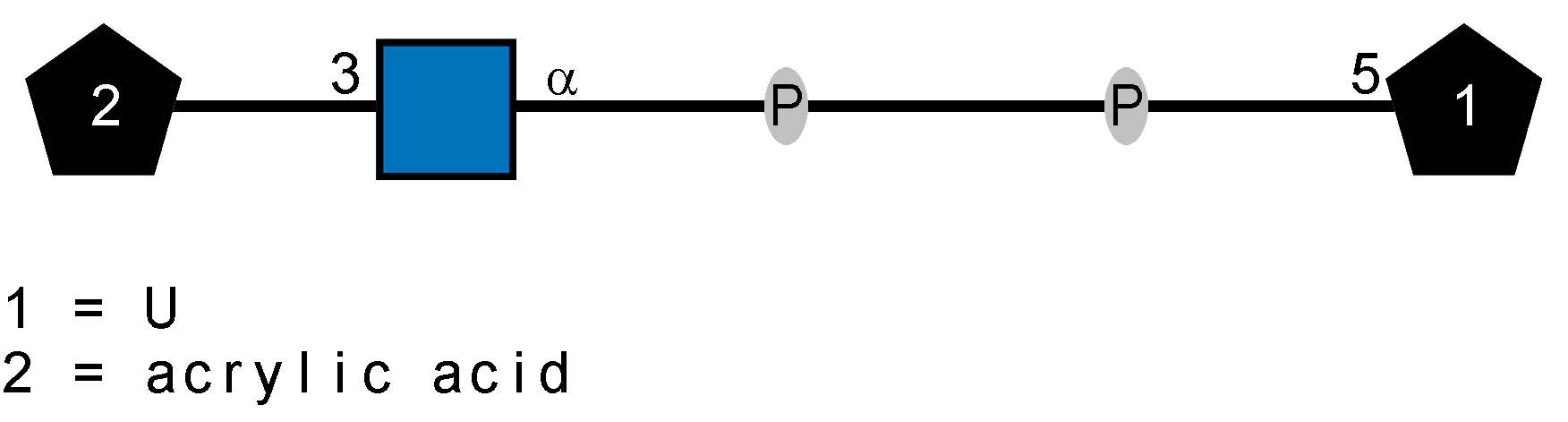
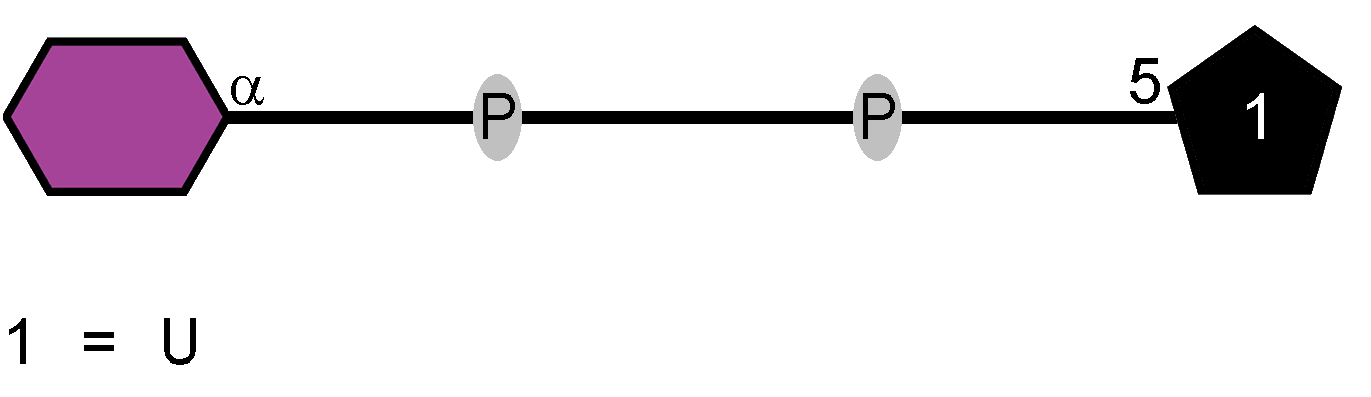
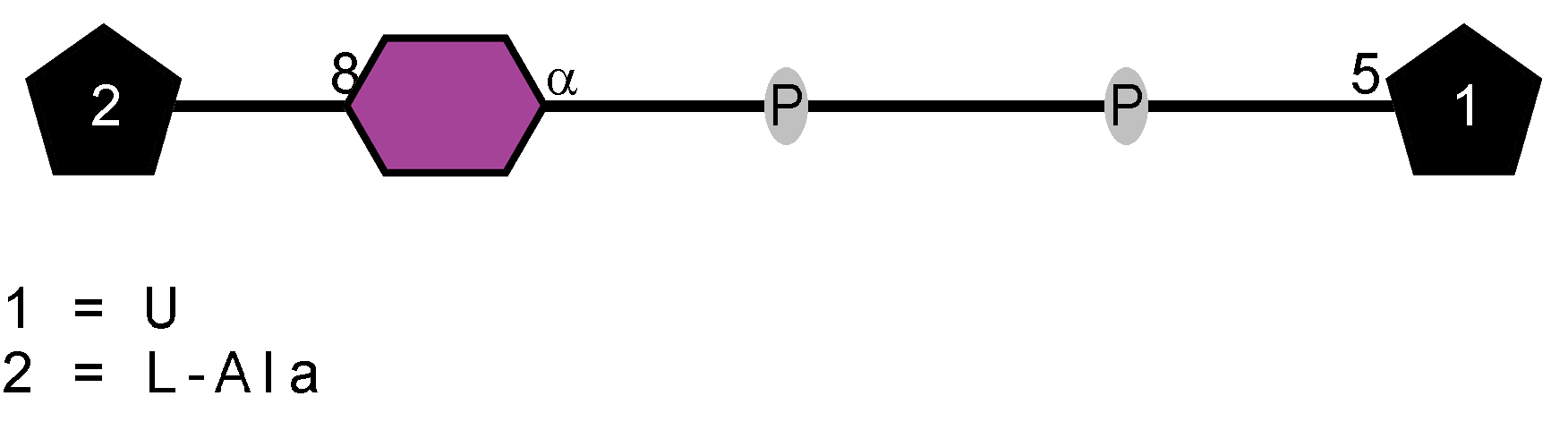
LOS
N-glycan
Capsular polysaccharide
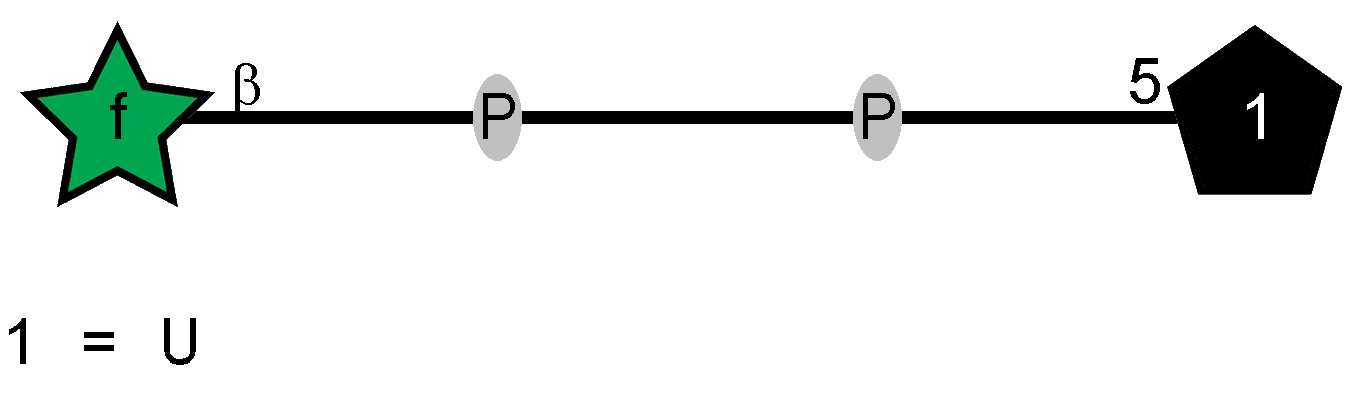
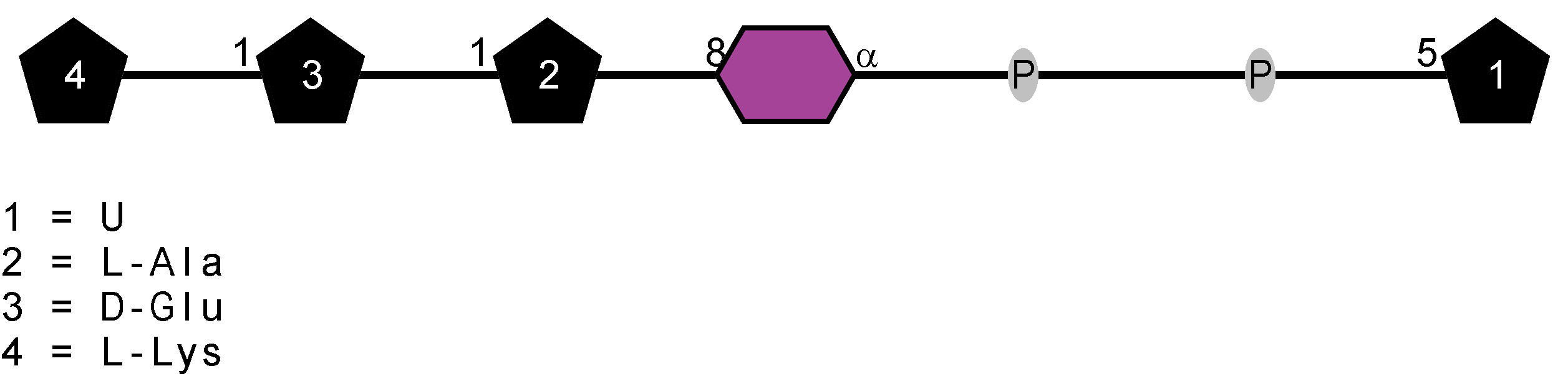
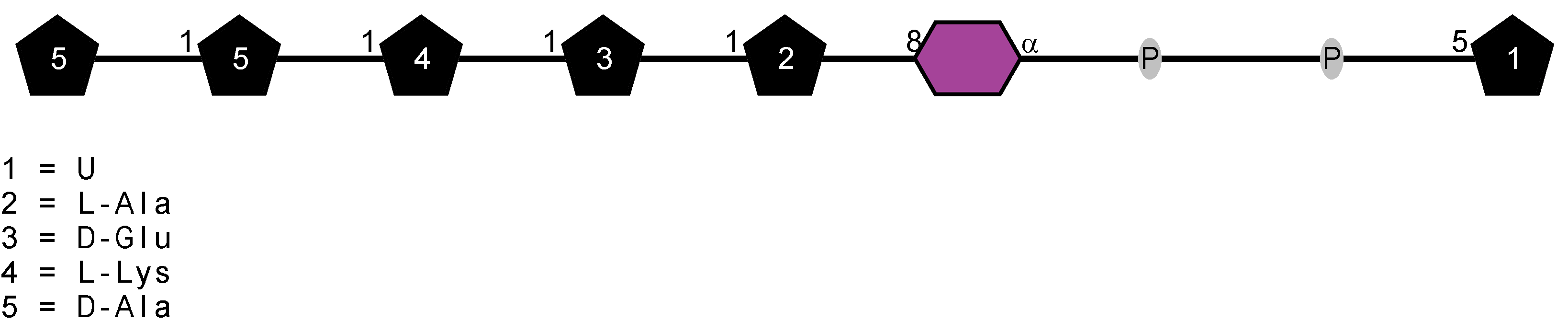
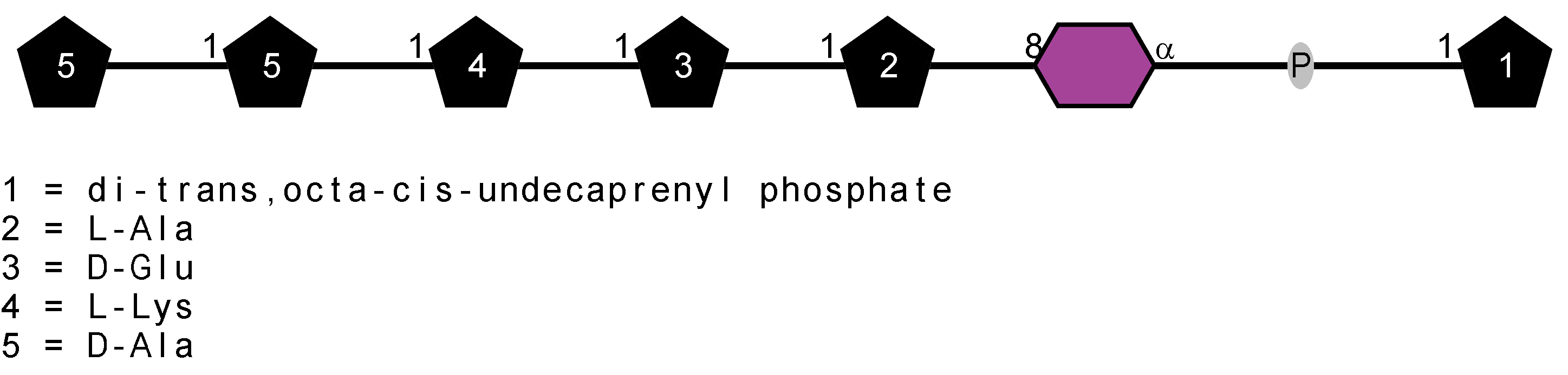
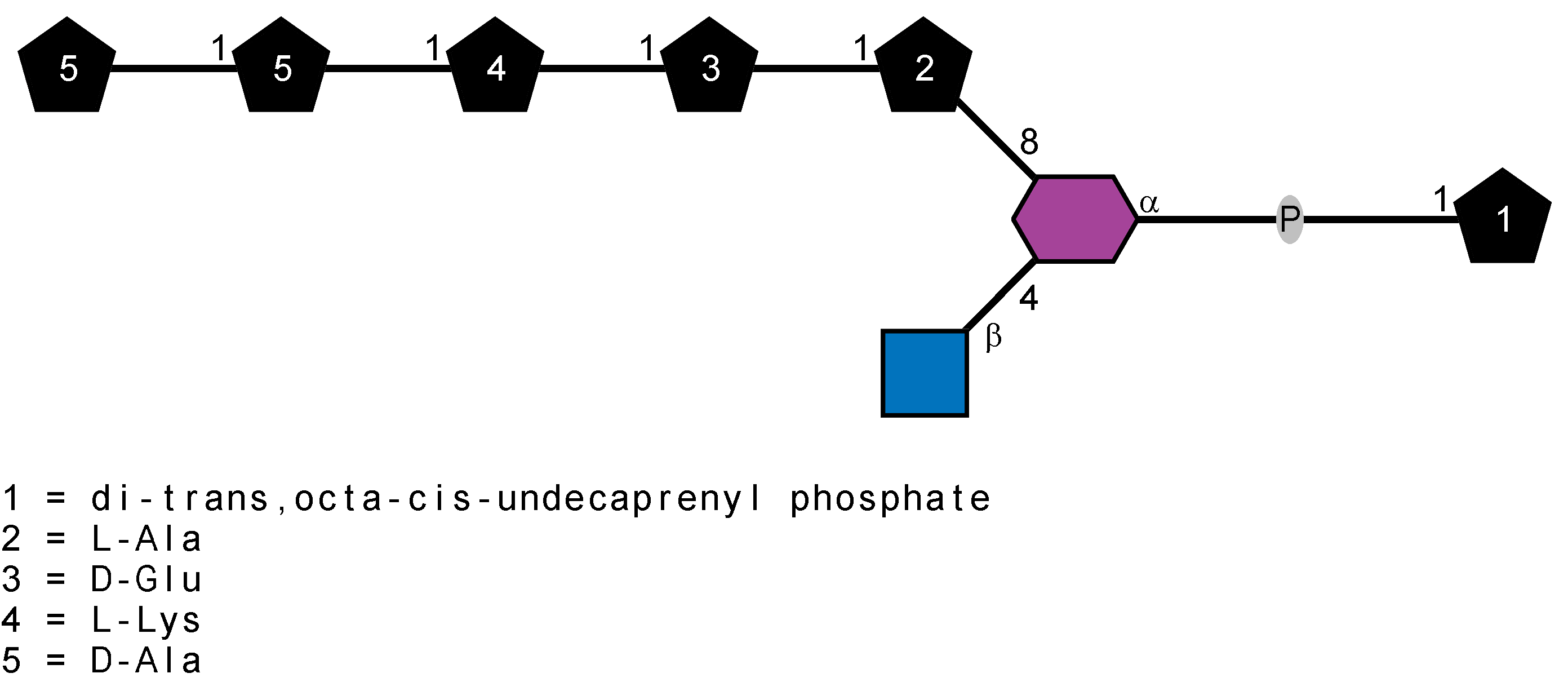
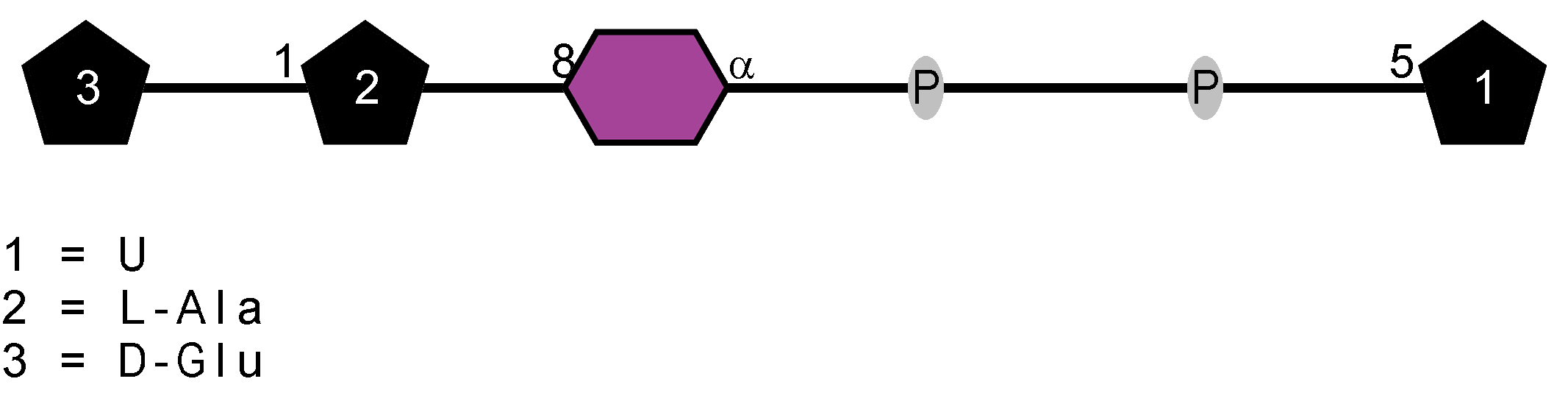
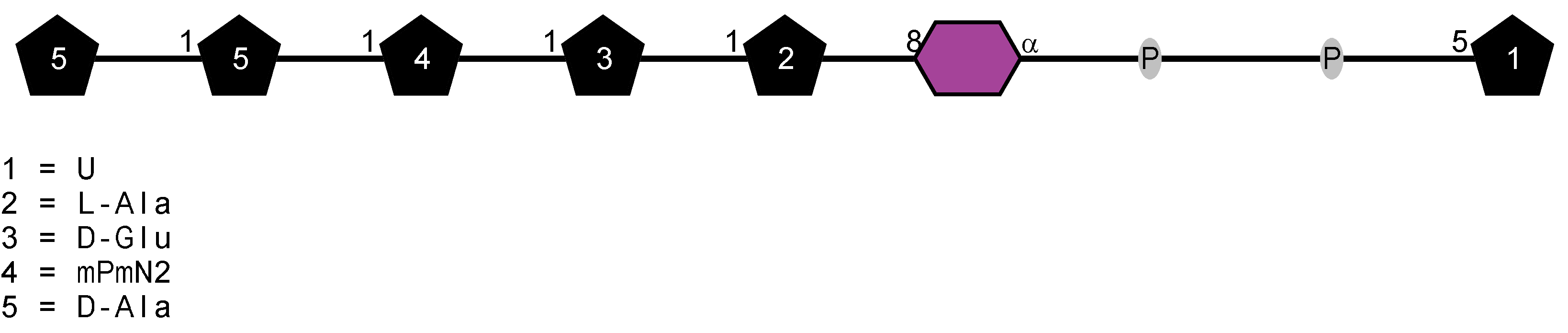
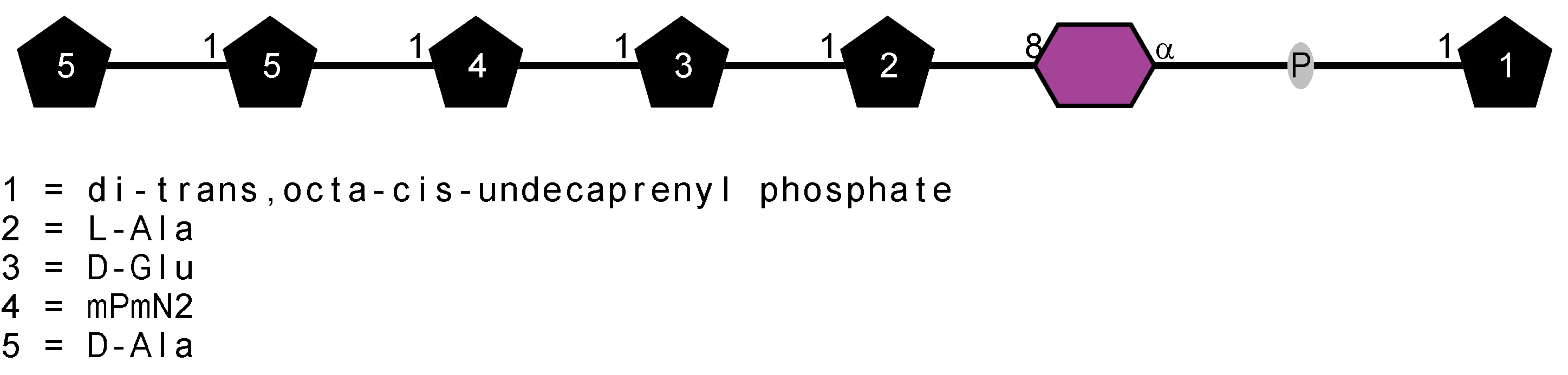
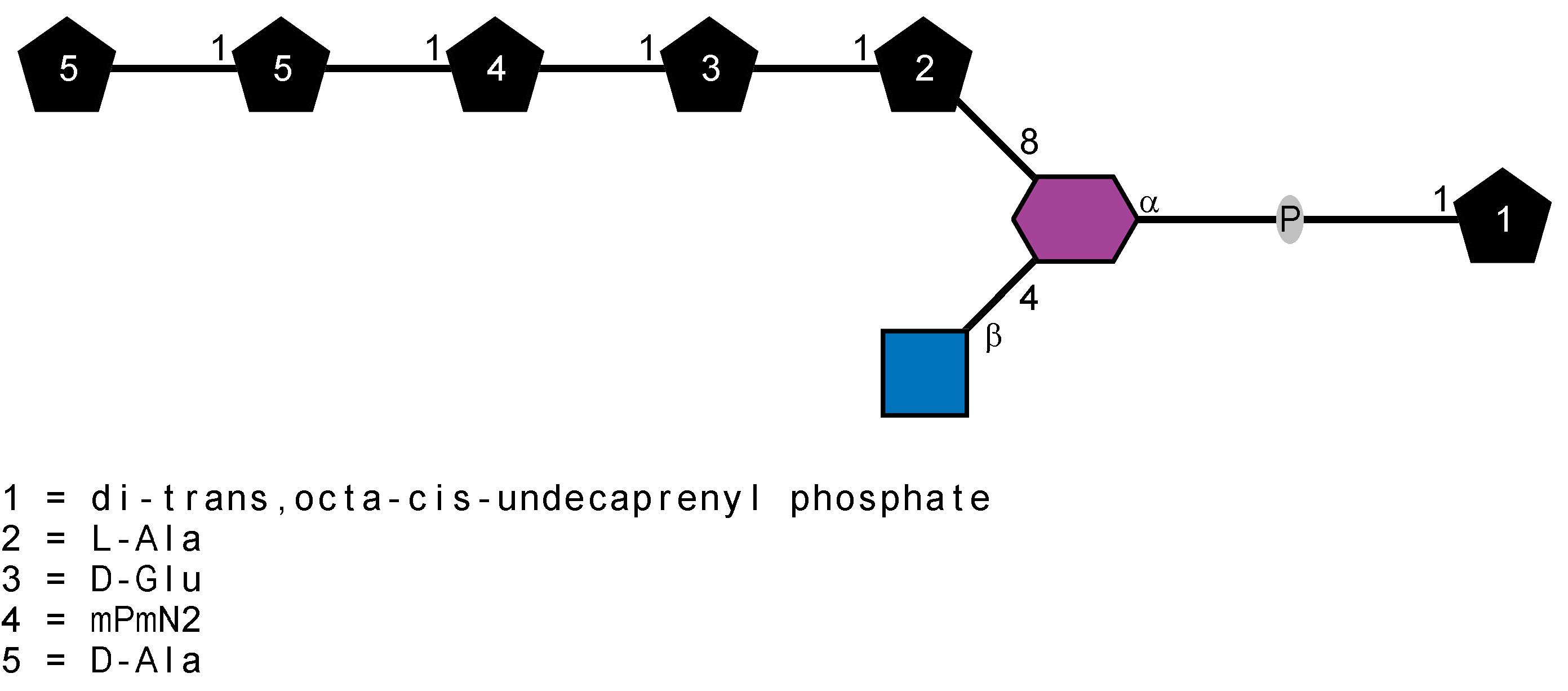
Peptidoglycan
Flagellin O-glycan
| MicroGlycoDB Gene ID | Gene No. | Gene Name | NOTES - phase variation etc |
|---|---|---|---|
| microglycodb_gene_00071 | Cj1132c | Unknown, conserved hypothetical protein | |
| microglycodb_gene_00072 | Cj1133 | waaC | Heptosyltransferase I |
| microglycodb_gene_00073 | Cj1134 | htrB | Lipid A biosynthesis acyltransferase |
| microglycodb_gene_00074 | Cj1135 | Two-domain glucosyltransferase: β-1,4-glucosyltransferase (N-terminal domain) and β-1,2-glucosyltransferase (C-terminal domain) In some C. jejuni strains Cj1135 contains a frameshift mutation that disrupts the C-terminal domain; those strains are lacking Glc on HepII | |
| microglycodb_gene_00075 | Cj1136 | β-1,3-Galactosyltransferase | |
| microglycodb_gene_00076 | Cj1137c | Glycosyltransferase | |
| microglycodb_gene_00077 | Cj1138 | Glycosyltransferase | |
| microglycodb_gene_00078 | Cj1139c | wlaN | Terminal Gal attached to β-1,4 GalNAc, a wlaN mutant is no longer able to bind cholera toxin (CT), phase variable |
| microglycodb_gene_00079 | Cj1140 | cstIII | Attaches to GalII, N-acetylneuraminic acid or sialic acid (cgtA/neuA1 (Cj1143), neuB1(Cj1141) and neuC1(Cj1142) are required for sialic acid biosynthesis) |
| microglycodb_gene_00080 | Cj1141 | neuB1 | Sialic acid synthase (N-acetylneuraminic acid synthetase) |
| microglycodb_gene_00081 | Cj1142 | neuC1 | UDP-N-acetylglucosamine 2-epimerase / N-acetyl-mannosamine synthase |
| microglycodb_gene_00082 | Cj1143 | cgtA | β-1,4-N-Acetylgalactosamine attached to GalII, and CMP-Neu5Ac synthesis by the C-terminal domain of Cj1143 |
| microglycodb_gene_00083 | Cj1144c | Unknown, phase variable | |
| microglycodb_gene_00084 | Cj1146c | waaV | Unknown, putative Glc-transferase |
| microglycodb_gene_00085 | Cj1148 | waaF | Heptosyltransferase II |
| microglycodb_gene_00086 | Cj1149c | gmhA | Sedoheptulose 7-phosphate isomerase (redundant with Cj1424c) |
| microglycodb_gene_00087 | Cj1150c | hldE | Bifunctional: D-glycero-b-D-mannoheptose 7-phosphate kinase / D-glycero-b-D-mannoheptose 1-phosphate adenylyltransferase |
| microglycodb_gene_00088 | Cj1151c | hldD | ADP-glyceromanno-heptose 6-epimerase |
| microglycodb_gene_00089 | Cj1152c | gmhB | D-glycero-D-mannoheptose 1,7-bisphosphate phosphatase |
| microglycodb_gene_00090 | Cj1293 | pseB | UDP-GlcNAc-specific C4,6 dehydratase / C5 epimerase |
| microglycodb_gene_00091 | Cj1294 | pseC | C4 aminotransferase specific for PseB product |
| microglycodb_gene_00092 | Cj1295 | Conserved hypothetical protein | |
| microglycodb_gene_00093 | Cj1296 | Hypothetical protein | |
| microglycodb_gene_00094 | Cj1297 | Hypothetical protein | |
| microglycodb_gene_00095 | Cj1298 | Putative N-acetyltransferase | |
| microglycodb_gene_00096 | Cj1299 | acpP2 | Putative acyl carrier protein |
| microglycodb_gene_00097 | Cj1300 | Putative SAM domain containing methyltransferase | |
| microglycodb_gene_00098 | Cj1301 | Hypothetical protein | |
| microglycodb_gene_00099 | Cj1302 | Putative HAD-superfamily phosphatase, subfamily IIIC | |
| microglycodb_gene_00100 | Cj1303 | fabH2 | Putative 3-oxoacyl-[acyl-carrier-protein] synthase |
| microglycodb_gene_00101 | Cj1304 | acpP3 | Putative 3-oxoacyl-[acyl-carrier-protein] synthase |
| microglycodb_gene_00102 | Cj1305c | Hypothetical protein | |
| microglycodb_gene_00103 | Cj1306c | Hypothetical protein | |
| microglycodb_gene_00104 | Cj1307 | Putative amino acid activating enzyme | |
| microglycodb_gene_00105 | Cj1308 | Putative acyl carrier protein | |
| microglycodb_gene_00106 | Cj1309c | Hypothetical protein | |
| microglycodb_gene_00107 | Cj1310c | Hypothetical protein | |
| microglycodb_gene_00108 | Cj1311 | pseF | Putative acylneuraminate cytidylyltransferase |
| microglycodb_gene_00109 | Cj1312 | pseG | Nucleotidase specific for PseC product, UDP-4-amino4,6-dideoxy-beta-L-AltNAc |
| microglycodb_gene_00110 | Cj1313 | pseH | N-acetyltransferase specific for PseC product, UDP-4-amino-4,6-dideoxy-beta-L-AltNAc |
| microglycodb_gene_00111 | Cj1314c | hisF | Imidazole glycerol phosphate synthase subunit |
| microglycodb_gene_00112 | Cj1315c | hisH | Imidazole glycerol phosphate synthase subunit |
| microglycodb_gene_00113 | Cj1316c | pseA | Pseudaminic acid biosynthesis PseA protein |
| microglycodb_gene_00114 | Cj1317 | pseI | Pse synthetase |
| microglycodb_gene_00115 | Cj1318 | maf1 | Motility accessory factor (function unknown) |
| microglycodb_gene_00116 | Cj1319 | Putative nucleotide sugar dehydratase | |
| microglycodb_gene_00117 | Cj1320 | Putative aminotransferase (degT family) | |
| microglycodb_gene_00118 | Cj1321 | Putative transferase | |
| microglycodb_gene_00119 | Cj1322 | Hypothetical protein | |
| microglycodb_gene_00120 | Cj1323 | Hypothetical protein | |
| microglycodb_gene_00121 | Cj1324 | Hypothetical protein | |
| microglycodb_gene_00122 | Cj1325 | Putative methyltransferase | |
| microglycodb_gene_00123 | Cj1327 | neuB2 | N-acetylneuraminic acid synthetase |
| microglycodb_gene_00124 | Cj1328 | neuC2 | Putative UDP-N-acetylglucosamine 2-epimerase |
| microglycodb_gene_00125 | Cj1329 | Putative sugar-phosphate nucleotide transferase | |
| microglycodb_gene_00126 | Cj1330 | Hypothetical protein | |
| microglycodb_gene_00127 | Cj1331 | ptmB | Acylneuraminate cytidylyltransferase (flagellin modification) |
| microglycodb_gene_00128 | Cj1332 | ptmA | Putative oxidoreductase (flagellin modification) |
| microglycodb_gene_00129 | Cj1333 | pseD | PseD protein |
| microglycodb_gene_00130 | Cj1334 | maf3 | Motility accessory factor (function unknown) |
| microglycodb_gene_00131 | Cj1335 | maf4 | Motility accessory factor (function unknown) |
| microglycodb_gene_00132 | Cj1337 | pseE | PseE protein |
| microglycodb_gene_00133 | Cj1338c | flaB | Flagellin subunit B |
| microglycodb_gene_00134 | Cj1339c | flaA | Flagellin subunit A |
| microglycodb_gene_00135 | Cj1340c | Conserved hypothetical protein | |
| microglycodb_gene_00136 | Cj1341c | maf6 | Motility accessory factor (function unknown) |
| microglycodb_gene_00137 | Cj1342c | maf7 | Motility accessory factor (function unknown) |
| microglycodb_gene_00138 | Cj1119c | pglG | Putative integral membrane protein, unknown function |
| microglycodb_gene_00139 | Cj1120c | pglF | UDP-N-acetyl-alpha-D-glucosamine C6 dehydratase |
| microglycodb_gene_00140 | Cj1121c | pglE | UDP-4-keto-6-deoxy-GlcNAc C4 aminotransferase |
| microglycodb_gene_00141 | Cj1122c | Putative integral membrane protein | |
| microglycodb_gene_00142 | Cj1123c | pglD | UDP-N-acetylbacillosamine N-acetyltransferase |
| microglycodb_gene_00143 | Cj1124c | pglC | Undecaprenyl phosphate N,N'-diacetylbacillosamine 1-phosphate transferase |
| microglycodb_gene_00144 | Cj1125c | pglA | Alpha-1,3-N-acetylgalactosaminyltransferase |
| microglycodb_gene_00145 | Cj1126c | pglB | Oligosaccharide transferase to N-glycosylate proteins and release FOS |
| microglycodb_gene_00146 | Cj1127c | pglJ | Alpha-1,4-N-acetylgalactosaminyltransferase |
| microglycodb_gene_00147 | Cj1128c | pgl | Beta-1,3-glucosyltransferase |
| microglycodb_gene_00148 | Cj1129c | pglH | Alpha-1,4-N-acetyl-galactosaminyltransferase, GlcNAc-polymerase |
| microglycodb_gene_00149 | Cj1130c | pglK | Flips LLO from cytoplasm into the periplasm |
| microglycodb_gene_00150 | Cj1131c | gne | UDP-GlcNAc / Glc 4-epimerase required for LOS, capsule and N-linked glycosylation |
| microglycodb_gene_00151 | Cj1413c | kpsS | Capsule polysaccharide modification protein |
| microglycodb_gene_00152 | Cj1414c | kpsC | Capsule polysaccharide modification protein |
| microglycodb_gene_00153 | Cj1415c | Cytidine diphosphoramidate kinase | |
| microglycodb_gene_00154 | Cj1416c | Cytidine triphosphate (CTP):phosphoglutamine cytidylyltransferase | |
| microglycodb_gene_00155 | Cj1417c | γ-glutamyl-CDP-amidate hydrolase | |
| microglycodb_gene_00156 | Cj1418c | L-glutamine kinase | |
| microglycodb_gene_00157 | Cj1419c | Phosphoramidate methyltransferase | |
| microglycodb_gene_00158 | Cj1420c | Phosphoramidate methyltransferase | |
| microglycodb_gene_00159 | Cj1421c | Adds phosphoramidate to C3 of the b-D-GalfNAc residue | |
| microglycodb_gene_00160 | Cj1422c | Adds phosphoramidate to C4 of D-glycero-a-L-gluco-heptopyranose | |
| microglycodb_gene_00161 | Cj1423c | hddC | D-glycero-D-manno-heptose 1-phosphate guanosyltransferase |
| microglycodb_gene_00162 | Cj1424c | gmhA2 | D-sedoheptulose-7-phosphate isomerase (redundant with Cj1149c) |
| microglycodb_gene_00163 | Cj1425c | hddA | D-glycero-D-manno-heptose 7-phosphate kinase |
| microglycodb_gene_00164 | Cj1426c | D-glycero-a-L-gluco-heptopyranose 6-O-methyltransferase | |
| microglycodb_gene_00165 | Cj1427c | (Putative GDP-6-R-D-glycero-D-lyxo-4-heptulose 4-reductase – 2013 Creuzenet; R = OH or OMe) | |
| microglycodb_gene_00166 | Cj1428c | (Putative GDP-6-R-L-xylo-4-heptulose 4-reductase – 2013 Creuzenet; R = OH or OMe) | |
| microglycodb_gene_00167 | Cj1429c | Hypothetical protein | |
| microglycodb_gene_00168 | Cj1430c | (putative GDP-6-R-D-glycero-D-lyxo-4-heptulose 3,5-epimerase – 2013 Creuzenet; R = OH or OMe) | |
| microglycodb_gene_00169 | Cj1431c | hddC | Capsular polysaccharide heptosyltransferase; (GDP-heptosyltransferase – Karlyshev) |
| microglycodb_gene_00170 | Cj1432c | Putative sugar transferase | |
| microglycodb_gene_00171 | Cj1433c | Hypothetical protein | |
| microglycodb_gene_00172 | Cj1434c | Putative sugar transferase | |
| microglycodb_gene_00173 | Cj1435c | Putative phosphatase | |
| microglycodb_gene_00174 | Cj1436c | Aminotransferase | |
| microglycodb_gene_00175 | Cj1437c | Aminotransferase | |
| microglycodb_gene_00176 | Cj1438c | Putative sugar transferase | |
| microglycodb_gene_00177 | Cj1439c | glf | UDP-GalNAc mutase – Poulin 2010 JBC |
| microglycodb_gene_00178 | Cj1440c | Putative sugar transferase | |
| microglycodb_gene_00179 | Cj1441c | kfiD | UDP-glucose 6-dehydrogenase |
| microglycodb_gene_00180 | Cj1442c | Putative sugar transferase | |
| microglycodb_gene_00181 | Cj1443c | kpsF | Capsule polysaccharide export system |
| microglycodb_gene_00182 | Cj1444c | kpsD | Capsule polysaccharide export system periplasmic protein |
| microglycodb_gene_00183 | Cj1445c | kpsE | Capsule polysaccharide export system inner membrane protein |
| microglycodb_gene_00184 | Cj1447c | kpsT | Capsule polysaccharide export ATP-binding protein |
| microglycodb_gene_00185 | Cj1448c | kpsM | Capsule polysaccharide export system inner membrane protein |
| microglycodb_gene_00186 | Cj1039 | murG | UDP-diphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase, leads to the formation of Lipid II |
| microglycodb_gene_00187 | Cj1676 | murB | Catalyzes the reduction of UDP N-acetylglucosamine enolpyruvate to form UDP-N-acetylmuramate in peptidoglycan biosynthesis |
| microglycodb_gene_00188 | Cj1054c | murC | Catalyzes the formation of UDP-N-acetylmuramoyl-L-alanine from UDP-N-acetylmuramate and L-alanine in peptidoglycan synthesis |
| microglycodb_gene_00189 | Cj0432c | murD | DP-N-acetylmuramoylalanine--D-glutamate ligase |
| microglycodb_gene_00190 | Cj0433c | mraY | First step of the lipid cycle Reactions in the biosynthesis of the cell wall peptidoglycan, leads to the formation of Lipid I |
| microglycodb_gene_00191 | Cj0858c | murA | Enolpyruvyl to UDP-N-acetylglucosamine as a component of cell wall formation |
| microglycodb_gene_00192 | Cj1641 | murE | Involved in cell wall formation; peptidoglycan synthesis; cytoplasmic enzyme; catalyzes the addition of meso-diaminopimelic acid to the nucleotide precursor UDP-N-aceylmuramoyl-l-alanyl-d-glutamate |
| microglycodb_gene_00193 | Cj0795c | murF | UDP-N-acetylmuramoyl-tripeptide |
| microglycodb_gene_00194 | Cj0798c | ddl | D-alanine--D-alanine ligase |
| microglycodb_gene_00195 | Cj1652c | murI | Converts L-glutamate to D-glutamate, a component of peptidoglycan |
| microglycodb_gene_00196 | Cj1038 | pycA | Cell division / peptidoglycan biosynthesis protein |